Challenge detailsChallenge progress
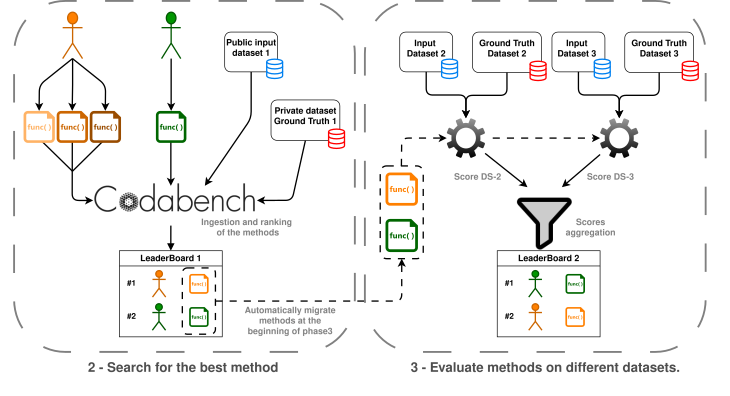
The challenge will be splited in three phases:
And in practice, how does it work?Participants will work in teams of 4 people. Everyone will work on their personal laptop and will have previously installed the necessary development environments for the challenge (Docker, R and associated packages). 1. Each team develops its solutions locally, on their personal computer. 2. Each team submits its solutions (its code or results) on the challenge platform, Codabench. 3. The code is executed on the platform using a standardized Docker environment. It results are submittes, these are compared with the results present in the platform (unknown of the teams). 4. It will be possible to make several submissions per team. The performance of each submission will be evaluated. 5. The best proposed solution (developed in R) from each team will then be transferred for the final evaluation of the challenge phase 3. 6. The Docker image will be available so that all teams can work in a standardized environment (in order to limit compatibility issues as much as possible). In summary, each team will develop its solution to the problem locally and then submit it to Codabench. Code execution and evaluation will be carried out via the standardized Docker environment. Submission platform: CodabenchThis challenge is hosted on Codabench, a data challenge platform: https:// Teams will submit their solutions on this platform. Solutions will be executed and evaluated there using a Docker image. Link to the challenge will be provided soon.
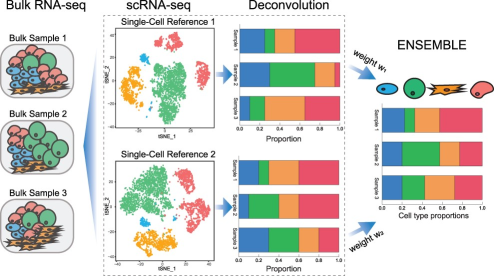
What is deconvolutionThe deconvolution of bulk RNAseq data estimates the proportion of different cell populations present in a sample, using gene expression profiles. This information can be used to study the cellular heterogeneity of tumors.
Overview of bulk RNAseq deconvolution using single-cell RNAseq reference. This figure is the Figure 1 from [1]. Datasets of the challenge
Different datasets will be used during the challenge:
References[1] Meichen Dong, Aatish Thennavan, Eugene Urrutia, Yun Li, Charles M Perou, Fei Zou, Yuchao Jiang, SCDC: bulk gene expression deconvolution by multiple single-cell RNA sequencing references, Briefings in Bioinformatics, Volume 22, Issue 1, January 2021, Pages 416–427, https://doi.org/10.1093/bib/bbz166 |